If haplotype A is seen x times in a sample of n haplotypes, pA = x / n. There has been a tendency in the literature arising from Holland and Parsons (ref. 1) to regard this quantity as being normally distributed and to acknowledge sampling variation with a 100(1 - メ)% confidence interval.
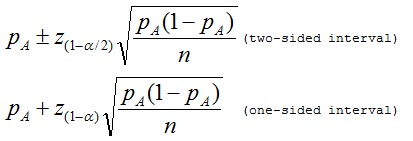
However, the actual sampling distribution is binomial. An exact confidence interval based on the binomial distribution was described by Clopper and Pearson (ref. 2).
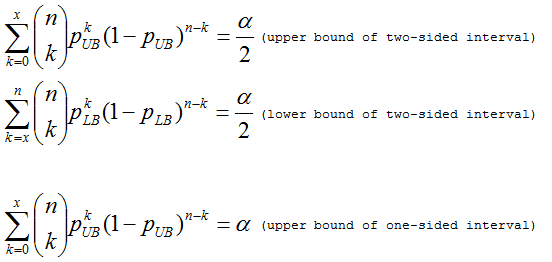
ySTRmanager provides an upper 95% one-sided confidence limit pUB (メ = 0.05) as an estimated frequency for a particular haplotype. If the sample does not contain any copies of A, then the above equation gives pUB = 1 - 0.051/n and this is approximately 3/n once n is 100 or more.
Exact binomial confidence intervals (pUB and pLB) can be obtained from MS Excel invoking "BETAINV" function and in R or S-Plus by the commmand "qbeta". They are also calculated in a web-site (http://statpages.org/confint.html).
| Two-sided interval | One-sided interval | ||
| Upper bound (pUB) | Lower bound (pLB) | Upper bound (pUB) | |
| MS Excel | BETAINV(1-メ/2,x+1,n-x) | BETAINV(メ/2,x,n-x+1) | BETAINV(1-メ,x+1,n-x) |
| R or S-plus | qbeta(1-メ/2,x+1,n-x) | qbeta(メ/2,x,n-x+1) | qbeta(1-メ,x+1,n-x) |
x is the number of matches in a particular population (or database).
n is the size of population (or database).
Reference
- Holland MM, Parsons TJ. Mitochondrial DNA sequence analysis - validation and use for forensic case work. Forensic Sci Rev 1999;11:21-50.
- Clopper CJ, Pearson ES. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika 1934;26(4):404-13.
- Buckleton JS, Krawczak M, Weir BS. The interpretation of lineage markers in forensic DNA testing. Forensic Sci Int Genet 2011;5(2):78-83.